using GMT
import Random
Random.seed!(1234);
x = -50.0:50;
y = x / 50 .+ 3 .+ 0.25 * rand(length(x));
# Add 3 outliers
y[[30,50,60]] = [4,-3,6];
viz(x, y, figsize=(15, 5))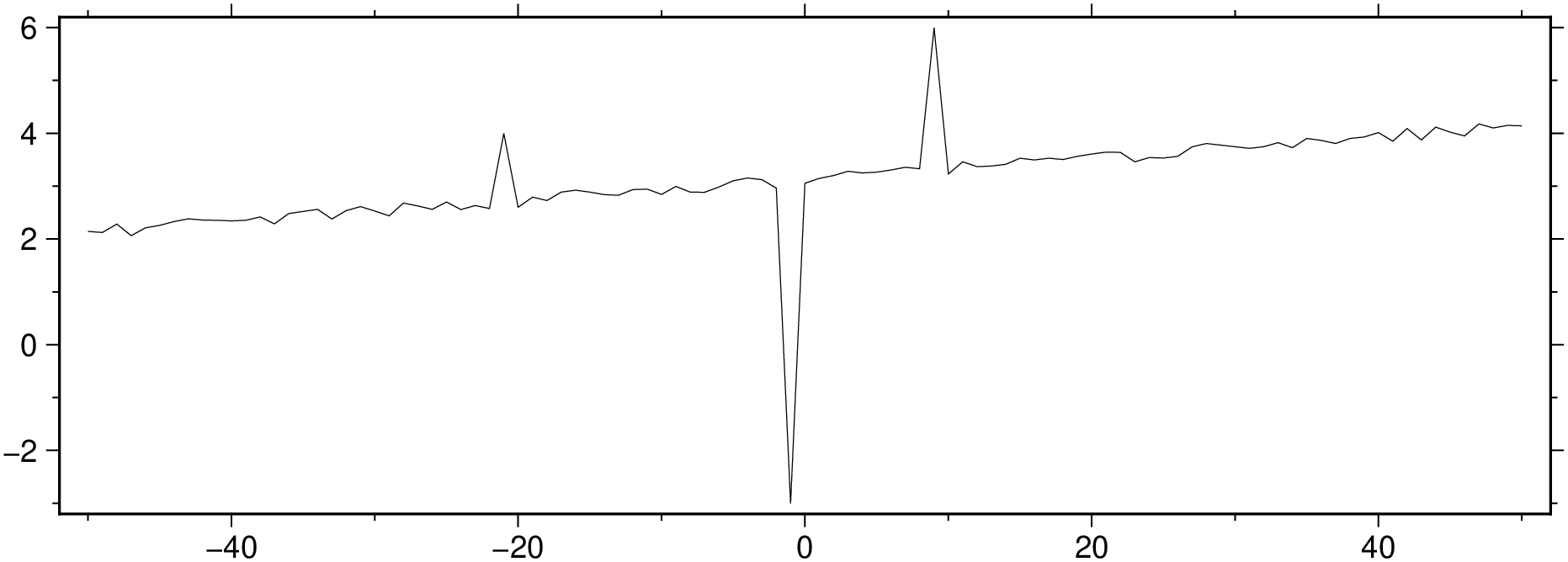
The examples in this tutorial show some ways in which we can easily remove the outliers of a time series.
Indentify the outliers positions with the help of the MAD estimator (MAD = median absolute deviation = 1.4826 * median(|xi−median(x)|))
The outliers positions are at:

Filter out the outliers with a median filter.
But what if we want to locale the outliers?
We can use the median filter but this time we ask filter1d do return the residues. We do that by setting the highpass option.
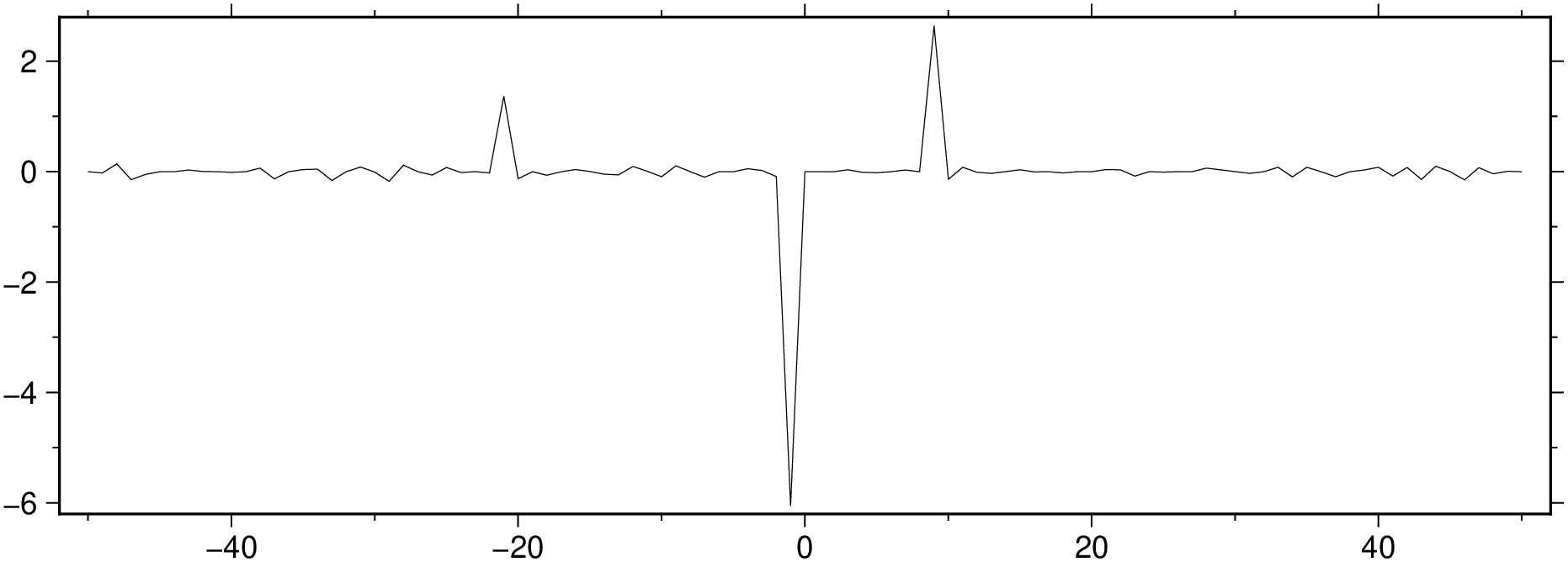
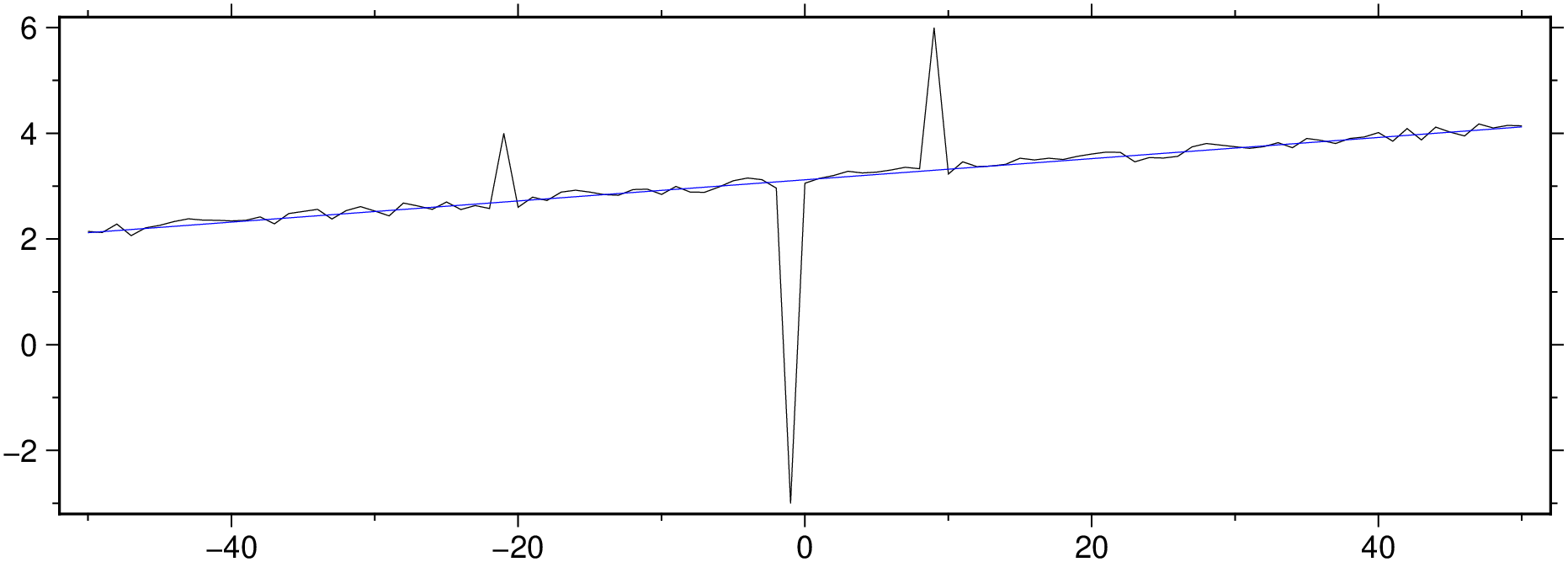
Fit a linear model and analyze the residues. The rin the out=:xmr option tells to output the residues too.
D = trend1d([x y], model=(polynome=1), out=:xmr)
BoundingBox: [-50.0, 50.0, 2.1219925238508743, 4.121792763622711, -6.101894641339074, 2.698125334683742]
101×3 GMTdataset{Float64, 2}
Row │ x model residues
─────┼──────────────────────────────
1 │ -50.0 2.12199 0.022973
2 │ -49.0 2.14199 -0.019167
3 │ -48.0 2.16199 0.121045
4 │ -47.0 2.18199 -0.118259
5 │ -46.0 2.20198 0.00810421
6 │ -45.0 2.22198 0.0379079
7 │ -44.0 2.24198 0.0879249
8 │ -43.0 2.26198 0.119807
⋮ │ ⋮ ⋮ ⋮
Show the residues
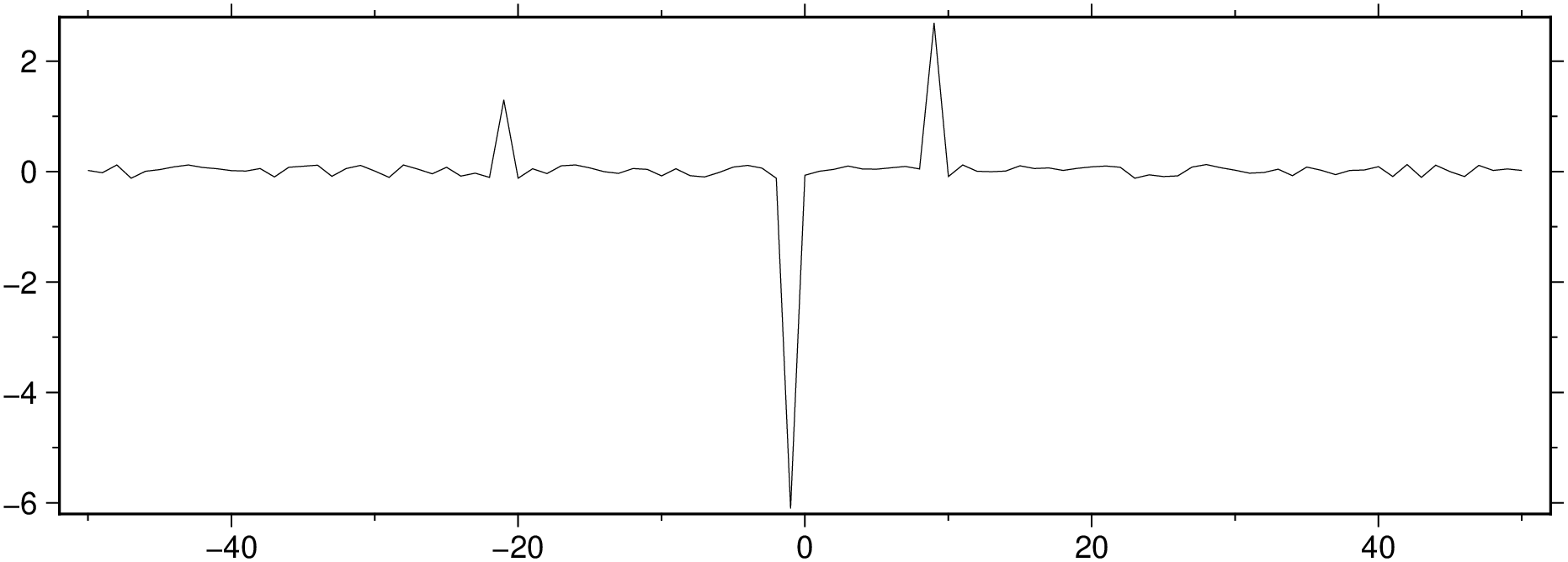
Random.seed!(1234);
x = -50.0:50;
y = x / 50 .+ 3 .+ 0.25 * rand(length(x));
y[[30,50,60]] = [4,-3,6];
D = trend1d([x y], model=(polynome=1), out=:xmr)
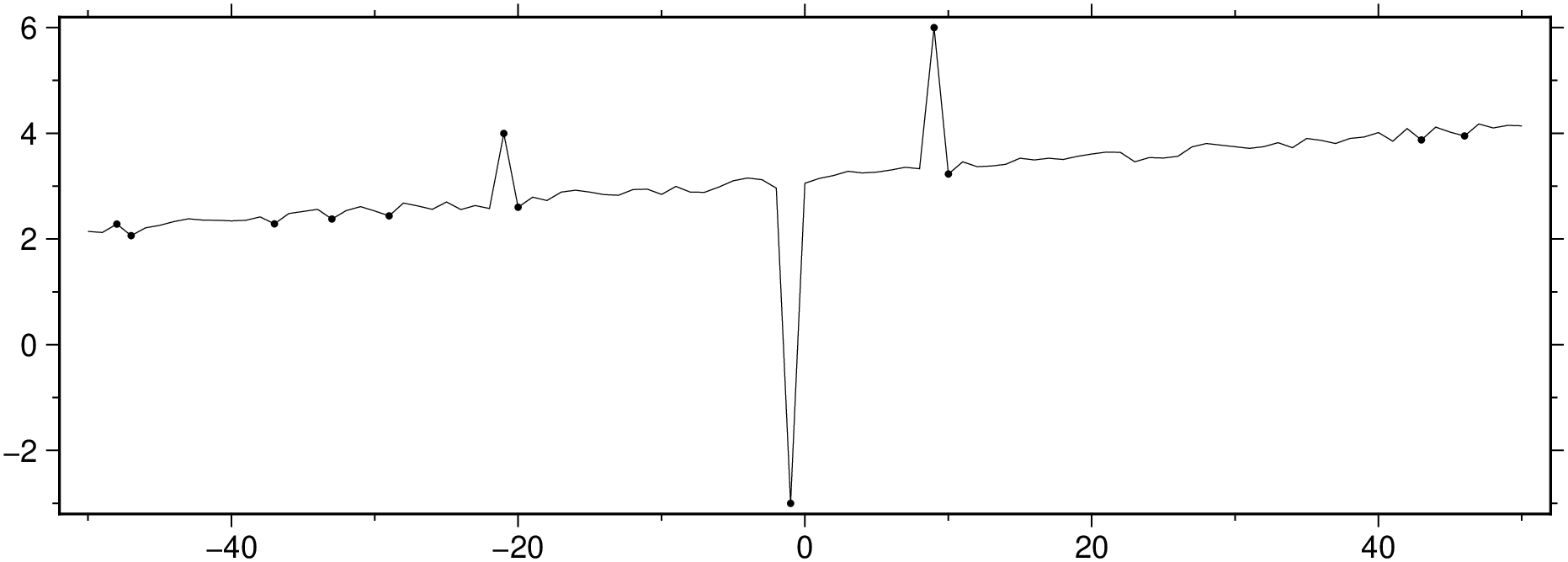
mad_res, med_res = mad(D[:,3])
zi = (D[:,3] .- med_res) ./ mad_res;
ind = abs.(zi) .> 3 * mad_res;
viz([x y], plot=(data=[x[ind] y[ind]], marker=:point), figsize=(15, 5))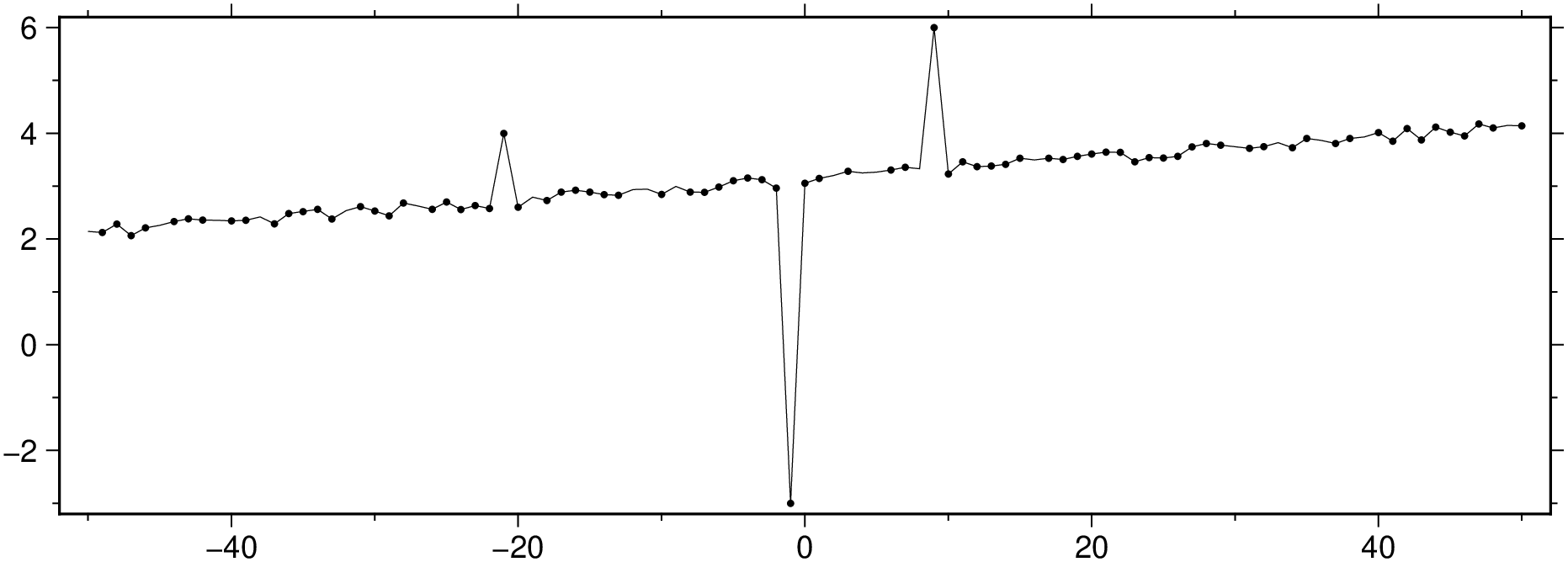
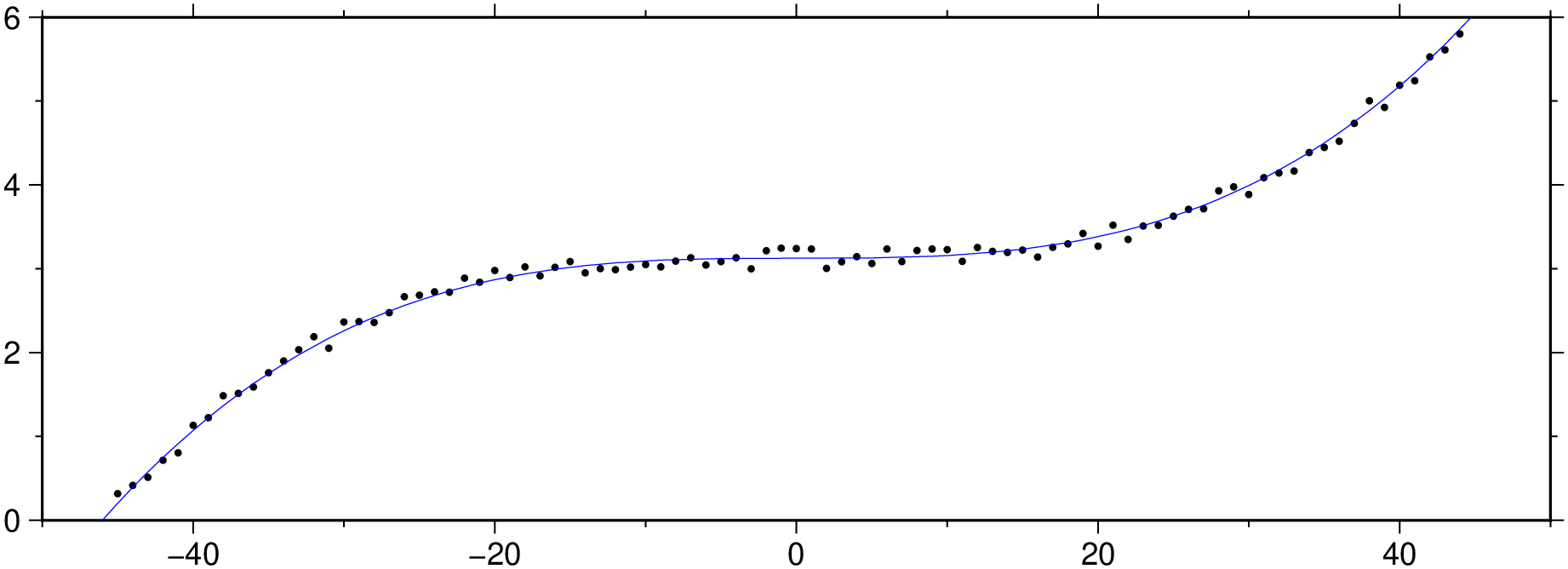
Let’s replace the outliers by NaNs.
And now replace the NaNs by interpolated values.
Polute it at positions [15, 40, 90].
Compute and show the residues.
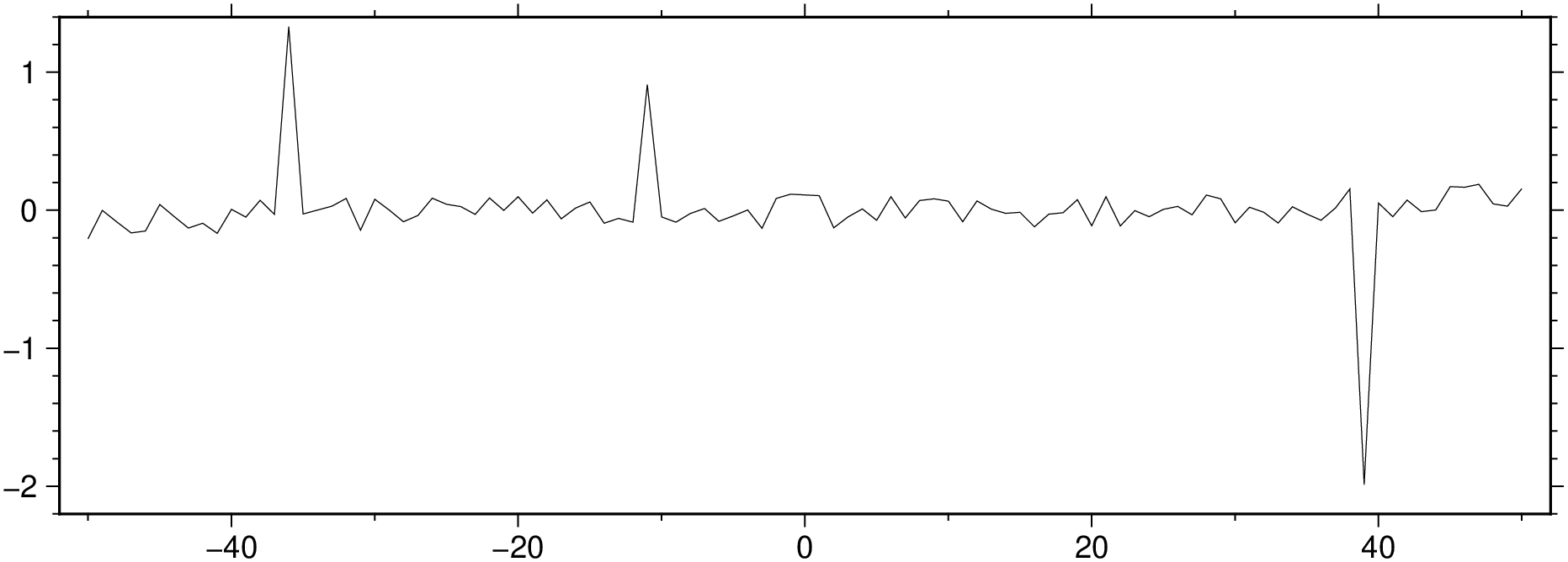
And now find the outliers positions and confirm that we obtain the locations used at the polution step.